Introduction
Recent Reviews
Safinya, C. R.; Deek, J.; Beck, R.; Jones, J. B.; Leal, C.; Ewert, K. K.; Li, Y.: Liquid crystal assemblies in biologically inspired systems. Liq. Cryst. 2013, 40, 1748-1758. DOI: 10.1080/02678292.2013.846422. ![]()
Safinya, C. R.; Raviv, U.; Needleman, D. J.; Zidovska, A.; Choi, M. C.; Ojeda-Lopez, M. A.; Ewert, K. K.; Li, Y.; Miller, H. P.; Quispe, J.; Carragher, B.; Potter, C. S.; Kim, M. W.; Feinstein, S. C.; Wilson, L.: Nanoscale Assembly in Biological Systems: From Neuronal Cytoskeletal Proteins to Curvature Stabilizing Lipids. Adv. Mater. 2011, 23, 2260-2270. DOI: 10.1002/adma.201004647. ![]()
Introduction and Motivation
The cytoskeleton is involved in a range of cell functions including cell attachment, cell division, mechanical stability, cell locomotion, intracellular trafficking and signal transduction. The main components of the cytoskeleton are three negatively charged filamentous proteins: filamentous actin, with a diameter of 8.5 nm; the family of intermediate filaments, ≈10 nm in diameter; and microtubules, 25 nm in diameter. These filamentous proteins self-assemble to form many different structures within the cell.
To a significant degree, our current understanding of biology is based on knowledge of the structures of biological molecules, which can provide direct information relating to function. With the emerging proteomics era, the biosciences now focus even more on elucidating the structures and functions of a large number of interacting proteins. Cellular activity often is a result of protein–protein and protein–nucleic acid interactions, which may lead to the formation of assemblies of biomolecules for distinct functions. Examples include the process of DNA condensation during the cell cycle and the formation of bundles and networks of cytoskeletal proteins for various purposes. Consequently, an important goal in biophysics is to understand the interactions leading to supramolecular structures of cytoskeletal proteins and associated biomolecules, and the elucidation of the roles of these structures in cell functions. We use synchrotron x-ray diffraction, electron microscopy, and various techniques of optical imaging to study the structure of these supramolecular assemblies on length scales from sub-nanometer to micrometer, from single filament to bundle arrangement. Purified proteins are combined in vitro to form systems designed to model the conditions faced by interacting proteins in vivo.
The current focus of our research is on understanding the structural nature of the cytoskeleton of nerve cell processes (axons and dendrites), which are comprised of a rich and complex variety of partially ordered bundles and networks of interacting neurofilaments, microtubules (MTs), and filamentous actin. The nature of these assembled protein structures, the interactions leading to them, and the correlations of structure and biological function remain poorly understood. We are studying nanoscale assembly in model systems comprised of reconstituted neurofilaments (NFs, purified from bovine spinal chord) and microtubule (MT)/microtubule-associated-protein (MAP) complexes. The long-range goals of this work are to identify parameters which control the forces leading to stable NF/MT networks and bundles, clarify their roles, and elucidate the biological functions of the hierarchical structures in neuronal processes.
The Structure of Actin Bundles
Introduction: Actin and Actin bundles

Figure 1. Confocal micrograph of mouse
fibroblast cells fluorescently stained to view
the actin cytoskeleton [1]. Note stress fibers
(broken arrow) and radial bundles of lamello-
podia (arrow). Scale bar = 10 μm.
Fig. 1 shows a laser scanning confocal microscope image of the actin cytoskeleton in mouse fibroblast cells, which highlights the importance and abundance of actin in cells. The actin cytoskeleton provides a structural framework for the mechanical stability of eukaryotic cells and is involved in functions including cell adhesion, motility, and division. Actin is found both in a monomeric globular (G) actin state and as polymerized filamentous actin (F-actin). The F-actin filament consists of a helical arrangement of G-actin sub-units. One repeat consists of 28 subunits over a length of 72 nm. Bundles, comprised of closely packed parallel strands of F-actin, and networks, containing F-actin crisscrossed at some large angle, form the most common supramolecular structures in cells. Interactions between F-actin and distinct actin crosslinking proteins may lead to two-dimensional (2D) networks and bundles of F-actin interacting with the plasma membrane to determine cell shape, or 3D networks of F-actin imparting gel-like properties to the cytosol.
In Figure 1, actin bundles traversing the length of a cell near the center of the image can be seen (broken arrow). These bundles are components of stress fibers, which end in focal adhesion spots responsible for cell adhesion, e.g., to the extracellular matrix in vivo. Radial bundles of lamellopodia in membrane-protrusions (Fig. 1, solid arrow) are also associated with adhesion complexes. An improved understanding of actin bundles, which may result from purely electrostatic interactions or crosslinking of F-actin by specific, actin-binding proteins, is a main goal of our work.

Figure 2. High resolution TEM image of two
negatively stained actin filaments and schematic
representation of an actin filament. Note how
the arrangement of globular protein
sub-units
(G-actin) leads to formation of the
overall helical
fiber structure.
Figure 2 shows a high resolution TEM image of two negatively stained actin filaments together with a schematic representation of the structure. The image illustrates how the arrangement of globular G-actin sub-units results in the overall helical fiber structure.
Actin-α-Actinin network structure
We have studied the interactions and structures in supramolecular assemblies of actin under in vitro conditions. As part of these investigations, we have determined the structure of filamentous actin (F-actin) complexed with the actin crosslinking protein α-actinin [1]. Stress fibres, lammellapodia and filapodia all contain α-actinin in actin bundles. This protein binds and crosslinks actin filaments under the correct salt conditions. The resulting actin bundles spontaneously form a remarkable new type of biologically inspired polyelectrolyte network.
Three-dimensional laser scanning confocal microscopy (LSCM) experiments revealed a relatively rigid, frequently branching, three-dimensional network of bundles on the micrometer scale (Fig. 3). This is in contrast to the commonly observed network of single filaments observed in cells. The branching (bifurcation) of the bundles on this meso-scale is evident and leads to a well-defined mesh size of the order of the persistence length of F-actin [1].
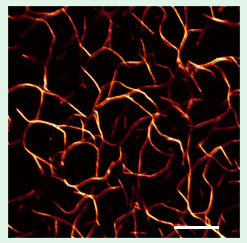
Figure 3. Confocal micrograph of F-actin
complexed with α-actinin in vitro (α-actinin:
G-actin=1:25): a network of bundles with
isotropic symmetry and a well-defined
mesh size [1]. Scale bar = 10 μm.
We probed the local, nanometer scale structural organization within the bundles using small angle x-ray diffraction (SAXRD) at the Stanford Synchrotron Radiation Laboratory (SSRL, beamlines 4-2 and 10-2) [1]. The experiments have revealed a structural transition from a loose network of filaments (at low crosslinker concentrations) to a disordered quasi-square lattice of actin fibers within bundles (at crosslinker concentrations near the physiological conditions). Experiments were done for different α-actinin:G-actin molar ratios (γ). A sharp increase in small angle x-ray scattering (SAXS) at γ = 1:50 (over, e.g., γ = 1:90) indicates the formation of macromolecular structures. This enhanced SAXS originates from the formation of a loose network of F-actin filaments (Fig. 2, left), where F-actin – F-actin correlations have not yet set in due to the low α-actinin concentration.
At γ ≤ 1:10, broad diffraction peaks observed in SAXRD indicate of the onset of F-actin correlations. The diffraction pattern reveals that the actin filaments are organized into a distorted (quasi-) square lattice within the bundles, with a unit cell dimension of approximately 30 nm (Fig. 4, right). The peak widths obtained from SAXRD are consistent with a model of inherent disorder within the bundles. Figure 4 (right) schematically shows the ladder-like structure within the bundles at a branching site.
Figure 4. Left: At low crosslinker
(red, ≈ 30
nm
length)
concentrations, SAXS reveals the
onset
of a loose network
of actin
filaments
(green). Right: At
higher crosslinker concentrations
diffraction peaks signal the onset
of F-actin bundles.
Shown in the
figure is a model of the structure
of an
F-actin bundle at a branching
site (derived from
synchrotron
SAXRD) with a disordered lattice with
quasi-square symmetry [1].
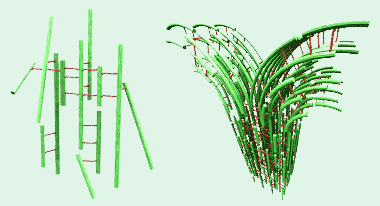
We have proposed that the bundling process proceeds in a zipper-like fashion between adjacent fibers. At a bifurcation point, an F-actin fiber will bend away from the main bundle and new fibers "zip" on, creating a branch.
[1] Pelletier, O.; Pokidysheva, E.; Hirst, L. S.; Bouxsein, N.; Li, Y.; Safinya, C. R.: Structure of actin cross-linked with alpha-actinin: A network of bundles. Phys. Rev. Lett. 2003, 91, 148102. ![]() (cover of Oct 3 issue)
(cover of Oct 3 issue)
Microtubules
Microtubules (MTs) have the largest diameter and stiffness of the three main components of the cellular cytoskeleton. They are essential for cell division, cell structure, and intracellular transport in eukaryotes. MTs are hollow cylinders of ~25 nm diameter, with a net negative charge. They are supramolecular polymers which self-assemble from αβ-tubulin protein heterodimers, as shown schematically in Figure 1. The tubulin dimers, whose structure has been determined at atomic detail [1], stack head to tail into protofilaments. These interact laterally, forming the MT wall [2].
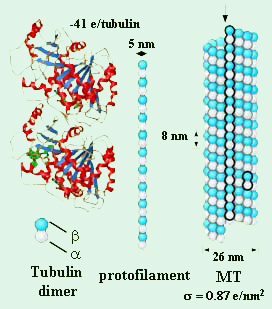
Figure 1. Structure of tubulin [6] and schematic depiction
of the assembly of tubulin heterodimers into micro-
tubules (MTs) .
Protofilaments form sheets at the ends of growing microtubules [3]. At the ends of shortening microtubules, protofilaments curl and peel apart from each other, eventually detaching from the microtubule as highly curved oligomers [4]. In contrast to this detailed structural knowledge, the forces and interactions that control MT dynamics are not well understood. There is no comprehensive model for the unique and complex manner of MT polymerization, called dynamic instability, in which MTs stochastically switch between growing and shortening phases. However, it is thought that the curvature of protofilaments and the interaction between protofilaments are two of the key microscopic determinants [5].
We purify tubulin from bovine brain in our lab or obtain it from our collaborator, Prof. Les Wilson. Most studies are performed using taxol-stabilized MTs.
[1] Lowe, J.; et al.: Refined structure of alpha beta-tubulin at 3.5 Å resolution. J. Mol. Biol. 2001, 313, 1045-1057.
[2] Li, H.; et al.: Microtubule structure at 8 Å resolution. Structure 2002, 10, 1317-1328.
[3] Chretien, D., Fuller, S. D.; Karsenti, E.: Structure of Growing Microtubule Ends: Two-Dimensional Sheets Close Into Tubes at Variable Rates. J. Cell Biol. 1995, 129, 1311-1328.
[4] Mandelkow, E. M.; Mandelkow, E.; Milligan, R. A.: Microtubule Dynamics and Microtubule Caps: A Time-Resolved Cryo-Electron Microscopy Study. J. Cell Biol. 1991, 114, 977-991.
[5] Desai, A.; Mitchison, T. J.: Microtubule polymerization dynamics. Annu. Rev. Cell Dev. Biol. 1997, 13, 83-117.
[6] Nogales, E.; Wolf, S. G.; Downing, K. H.: Structure of the alpha beta tubulin dimer by electron crystallography. Nature 1998, 391, 199-203.
Microtubule Buckling
In this section, we describe how we have used SAXS investigations of microtubule (MT) buckling under osmotic pressure to gain insight into the different mechanisms by which MT-associated proteins and MT stabilizing cancer drugs mechanically stabilize microtubules.
MTs, MAPs and chemotherapeutic drugs
MT polymerization is strongly affected by solution conditions, such as the ionic strength [1] and the presence of drugs and microtubule associated proteins (MAPs). Many anti-cancer drugs currently in use in clinical chemotherapy act by interfering with MT dynamics [2]. For example, taxol, which is widely used to treat breast and ovarian cancer, prevents cell division by binding to and stabilizing MTs [3]. However, the molecular mechanism by which taxol affects MTs is unclear. It has been proposed that taxol stabilizes MTs by either increasing the lateral interactions between protofilaments [4] or by locking the protofilaments in a straight conformation [5].
MAPs regulate the dynamic instability in vitro [6] and in cells [7]. A number of neurodegenerative diseases are associated with improper MAP–MT interactions [8]. Nontheless, the microscopic mechanism by which distinct MAPs affect MT dynamics is not well understood. It is unclear if MT-stabilizing MAPs function by crosslinking tubulin dimers within a protofilament or between protofilaments, or if they change the protofilament’s curvature. They could also cause a conformational change in tubulin or work by a completely different mechanisms [9].
Depletion–attraction and osmotic stress
The presence of inert macromolecules (e.g. PEG) can lead to an effective attraction between particles. This "depletion–attraction" force is well understood [10] and has been observed for DNA [11], actin [12], and MTs [13]. All of these are rod-like polymers which form bundles in the presence of inert macromolecules.
The centers of the inert macromolecules are excluded from a region around each rod due to steric repulsion. If these ‘excluded volumes’ around the rods are made to overlap with each other, the space available to the macromolecules increases, which increases the total entropy. This results in an effective attraction between the rods.
The depletion–attraction phenomenon can be used to create a powerful probe of intermolecular interactions if the induced attraction causes the macromolecules to phase separate from the rod-like particles [14]. In this method, a known osmotic pressure is applied by controlling the concentration of inert macromolecule, and the resulting spacing between particles is determined by x-ray scattering. This so-called osmotic stress technique has been used to study, e.g., the forces between viruses [15] and the interactions between DNA molecules [16,17]. As in these cases, the interaction between MTs is dominated by electrostatic double layer forces.
Osmotic stress on (hollow) MTs
Unlike most other rod-like biological polyelectrolytes such as actin or DNA, MTs are hollow. This leads to additional possible effects of applied osmotic pressure. If the added inert macromolecule is large enough to be excluded from the MT’s inside space, a pressure imbalance is created between the inside and outside of the MT. For a sufficiently large pressure difference, the MTs buckle to a nonspherical cross section. As the microtubules buckle, they also form bundles [13]. This is not surprising since the strength of depletion–attraction is highly dependent on the shape of the attracting particles [18, 19]. It can be orders of magnitude greater between flattened particles compared to rounded particles, i.e., between distorted and undistorted MTs.
The buckling of MTs is analogous to the buckling of a hollow tube with closed ends subjected to enormous hydrostatic pressure, such as a submarine in deep sea [13]. This deformation of tubes under pressure can be used to measure the radial mechanical properties of the tubes. Knowledge of MT mechanics can be used to gain insight into the molecular and mesoscopic interactions that underlie MT polymerization and depolymerization and, ultimately, their behavior and functions in vivo [20–24]. Specifially, the inter-protofilament interactions govern the -response of MTs to osmotic stress and can be probed (for the first time) using the osmotic stress–SAXRD method we have developed. The method thus allows direct investigation of the microscopic mode of action of MT stabilizing agents by using mechanical perturbations.
Microtubules buckle and bundle under osmotic stress
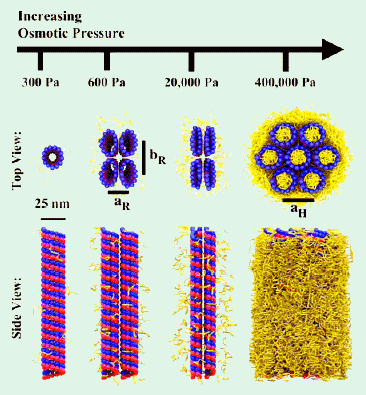
Figure 2. Summary of the effect of osmotic stress experiments
on MTs using 20 kDa PEG [25]. At low concentrations of PEG, the
MTs are undistorted and form a nematic. Above 0.4% PEG,
the MTs buckle to a noncircular cross section and form bundles
with rectangular symmetry. The MTs distort further as the
osmotic pressure increases. At 5% 20 kDa PEG (which approx-
imately equals c*, the polymer’s overlap concentration), the PEG
is forced inside the lumen of the MTs, and the MTs convert to
undistorted MTs in hexagonal bundles.
Our results [13,25] are summarized in Figure 2. With no or low concentration of added osmolytes, the added polymers have no effect on the MTs, but the MTs align due to steric interactions with each other. When the concentration of stressing polymer is increased, the MTs form bundles due to depletion–attraction. If the polymers do not enter the MT lumen, the MTs buckle to a noncircular cross section and pack into a lattice with rectangular symmetry. The MTs continue to distort as the osmotic pressure increases. This buckling and distortion and the corresponding bundling are reversible. If the polymer can enter the MT, because of its small size or at high concentrations of medium-size polymer, hexagonal bundles of undistorted MTs are formed by depletion–attraction. We have used the osmotic stress technique in this hexagonal bundle phase to measure the interactions between MTs, while studies of the rectangular bundle phase reveal interactions of protofilaments within the MTs.
Optical microscopy directly confirms the connection between formation of the rectangular bundle phase and exclusion of stressing polymer from the MT lumen. Rectangular bundles created with fluorescent 500 kDa dextran (Figure 3, 8% 500 kDa dextran) or small amounts of fluorescently labeled 20 kDa PEG (Figure 3, 1% 20 kDa PEG) appear dark in fluorescence. Thus, the dextran and 20 kDa PEG are excluded from the bundles. If now the concentration of unlabeled 20 kDa PEG is increased while keeping the concentration of fluorescently labeled 20 kDa PEG constant, the bundles move into the hexagonal phase. These bundles are not different from background in fluorescence images (Figure 3, 20% 20 kDa PEG), consistent with 20 kDa PEG entering the lumen of MTs as they form hexagonal bundles.
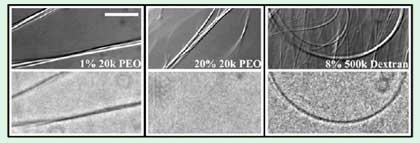
Figure 3. DIC microscopy (top) and corresponding fluorescence
microscopy (bottom) images of MTs with 1% 20 kDa PEG, 20% 20
kDa PEG, and 8% 500 kDa dextran (scale bar = 10 μm) [25]. All samples
contain 100 mM monovalent salt. Both PEG samples contain 0.1%
fluorescently labeled 20 kDa PEG. All of the dextran is fluorescently
labeled. The bundles formed with 1% 20 kDa PEG and 8% 500 kDa
dextran, which SAXD identifies as rectangular, appear dark in fluorescence
mode. This indicates that the polymers are excluded from these bundles. In
contrast, the fluorescence images of the (hexagonal) bundles formed
with 20% 20 kDa PEG are uniform, consistent with PEG entering
these bundles.
The effect of MAPs, salt and taxol on microtubule buckling and bundling
MTs in neurons are much more stable than MT in other cell types due to the action of MAPs [6,7]. The presence of neuronal MAPs strongly affects MT bundling in our experiments. SAXRD showed that even the presence of small amounts of MAPs greatly suppresses formation of rectangular bundles. MTs with ≈15% MAPs (i.e., 15% of the amount in vivo) do not display the rectangular bundle phase at any osmotic pressure, but the hexagonal bundle phase is still present with high concentrations of added 20 kDa PEG. No rectangular bundles are observed with 100% MAPs at physiological ionic strength. Thus, our osmotic stress–SAXRD technique provides the first demonstration that MAPs increase inter-protofilament interactions. This presumably happens by MAPs directly crosslinking protofilaments, but may also be due to a MAP-induced conformational change in tubulin.
In contrast to the effects of MAPs, which increase the pressure required to induce rectangular bundles, salt (KCl) decreases that pressure. This is not surprising, as observations from high resolution MT models along with detailed computer simulations [26] suggest that the inter-protofilament bonds are largely electrostatic in nature.
However, the presence of the chemotherapy drug taxol has no effect on the nematic to rectangular bundle phase boundary. It is surprising that taxol and MAPs have such different effects on the transition to the rectangular bundle phase, since both taxol and MAPs stabilize MTs against depolymerization. In fact, MAPs have a more modest effect on MT polymerization than the probed range of taxol concentrations (2.5 μM to 20 μM) [3]. Thus, our data suggests that taxol does not stabilize MTs by increasing the interaction between protofilaments. Rather, it acts by a different microscopic mechanism than MAPs. Our results support an earlier suggestion that taxol functions by preventing the straight-to-curved conformational change normally associated with GTP hydrolysis [5].
Conclusions and future work
Our work raises questions about the state of MTs in the environment found inside of cells. Cytoplasm is typically 20% protein by weight, and macromolecular crowding is believed to be significant at such high concentrations. This has important ramifications for cellular architecture and macromolecular interactions [27]. The osmotic pressure inside of cells can be MPas [28], orders of magnitude greater than the osmotic pressure required to buckle MTs composed of pure tubulin. Even if the rectangular phase of buckled MTs is suppressed inside cells by associated MAPs or because some of the osmotic stressing agents enter the MT lumen, the osmotic pressure is still high enough to form hexagonal bundles through depletion–attraction; in fact, close packed hexagonal bundles of MTs have been observed in stressed cells [29].
Having established the usefulness of the osmotic stress–SAXRD technique for measuring inter-protofilament interactions, we are now applying this probe of the inter-protofilament bond strength to other stabilizing agents, such as specific MAPs, other drugs, GMPCPP, as well as destabilizing agents, such as Ca2+ and low temperatures.
Other projects involve more detailed investigations of MT–MT interactions and how various MAPs alter them.
[1] Olmsted, J. B.; Borisy, G.G.: Ionic and nucleotide requirements for microtubule polymerization in vitro. Biochemistry 1975, 14, 2996-3005.
[2] Jordan, M. A.; Wilson, L.: Microtubules as a target for anticancer drugs. Nature Rev. Cancer 2004, 4, 253-265.
[3] Derry, W. B.; Wilson, L.; Jordan, M. A.: Substoichiometric binding of taxol suppresses microtubule dynamics. Biochemistry 1995, 34, 2203-2211.
[4] Amos, L. A.; Lowe, J.: How taxol stabilises microtubule structure. Chem. Biol. 1999, 6, R65-R69.
[5] Arnal, I.; Wade, R. H.: How does taxol stabilize microtubules? Curr. Biol. 1995, 5, 900-908.
[6] Panda, D.; et al.: Kinetic Stabilization of microtubule dynamics at steady state by tau and microtubule-binding domains of tau. Biochemistry 1995, 34, 1117-1127.
[7] Bunker, J. M.; et al.: Modulation of microtubule dynamics by tau in living cells: implications for development and neurodegeneration. Mol. Biol. Cell 2004, 15, 2720-2728.
[8] Lee, V. M. Y.; Goedert, M.; Trojanowski, J. Q.: Neurodegenerative taupathies. Annu. Rev. Neurosci. 2001, 24, 1121-1159.
[9] Cassimeris, L.; Spittle, C.: Regulation of microtubule-associated proteins. Int. Rev. Cytol. 2001, 210, 163-226.
[10] Asakura, S.; Oosawa, F.: Interaction between particles suspended in solutions of macromolecules. J. Polym. Sci. 1958, 33, 183-192.
[11] Bloomfield, V.: DNA Condensation. Curr. Opin. Struct. Biol. 1996, 6, 334-341.
[12] Hosek, M.; Tang, J. X.: Polymer-induced bundling of F-actin and the depletion force. Phys. Rev. E 2004, 69, 051907.
[13] Needleman, D. .J.; Ojeda-Lopez, M. A.; Raviv, U.; Ewert, K.; Jones, J. B.; Miller, H. P.; Wilson, L.; Safinya, C. R.: Synchrotron X-ray Diffraction Study of Microtubules Buckling and Bundling under Osmotic Stress: A Probe of Inter-Protofilament Interactions. Phys. Rev. Lett. 2004, 93, 198104.![]()
[14] Parsegian, V. A.; Rand, R. P.; Rau, D. C.: Macromolecules and water: probing with osmotic stress. Methods Enzymol. 1995, 259, 43-94.
[15] Millman, B. M.; et al.: Interrod forces in aqueous gels of tobacco mosaic virus. Biophys. J. 1984, 45, 551-556.
[16] Rau, R. C., Lee, B.; Parsegian, V. A.: Measurement of the repulsive force between polyelectrolyte molecules in ionic solution: hydration forces between parallel DNA double helices. Proc. Natl. Acad. Sci. U.S.A. 1984, 81, 2621-2625.
[17] Strey, H. H.; et al.: DNA-DNA interactions. Curr. Opin. Struct. Biol. 1998, 8, 309-313.
[18] Dogic, Z.; et al.: Isotropic-nematic phase transition in suspensions of filamentous virus and the neutral polymer dextran. Phys. Rev. E 2004, 69, 051702.
[19] Mason, T. G.: Osmotically driven shape-dependent colloidal separations. Phys. Rev. E 2002, 66, 060402.
[20] Stukalin, E. B.; Kolomeisky, A. B.: Simple growth models of rigid multifilament bioplymers. J. Chem. Phys. 2004, 121, 1097-1104.
[21] Mickey, B.; Howard, J.: Rigidity of microtubules is increased by stabilizing agents. J. Cell Biol. 1995, 130, 909-917.
[22] Chretien, D.; Fuller, S. D.: Microtubules switch occasionally into unfavorable configurations during elongation. J. Mol. Biol. 2000, 298, 663-676.
[23] Janosi, I. M.; Chretien, D.; Flyvbjerg, H.: Structural microtubule cap: stability, catastrophe, rescue, and third state. Biophys. J. 2002, 83, 1317-1330.
[24] Desai, A.; Mitchison, T. J.: Microtubule polymerization dynamics. Annu. Rev. Cell Dev. Biol. 1997, 13, 83-117.
[25] Needleman, D. J.; Ojeda-Lopez, M. A.; Raviv, U.; Ewert, K.; Miller, H. P.; Wilson, L.; Safinya, C. R.: Radial compression of microtubules and the mechanism of action of taxol and associated proteins. Biophys. J. 2005, 89, 3410-3423.![]()
[26] Sept, D.; Baker, N.A.; McCammon, J. A.: The physical basis of microtubule structure and stability. Protein Sci. 2003, 12, 2257-2261.
[27] Walter, H.; Brooks, D. E.: Phase separation in cytoplasm, due to macromolecular crowding, is the basis for microcompartmentation. FEBS Lett. 1995, 361, 135-139.
[28] Bray, D.: Cell Movements. 2001, New York: Garland.
[29] Rieder, C.; Bajer, A. S.: Heat-induced reversible hexagonal packing of spindle microtubules. J. Cell Biol. 1977, 74, 717-725.
Bundling and Higher Order Assembly of Microtubules by Counterions:
From Hexagonal Bundles to Living Necklaces
Cells tightly regulate the architecture of bundles of filamentous cytoskeletal proteins, giving rise to assemblies with distinct morphologies and physical properties. A similar control of the supramolecular organization of nanotubes and nanorods in synthetic materials is highly desirable. However, many of the principles that determine how macromolecular interactions lead to assemblies with defined morphologies remain unknown. For example, while a large number of key proteins critical to the overall stability of the axonal cytoskeleton have been identified through molecular biology and genetics studies, we still have very little knowledge of the supramolecular structures that are assembled when the cytoskeleton of the nerve cell is constructed and dynamically repaired throughout life.
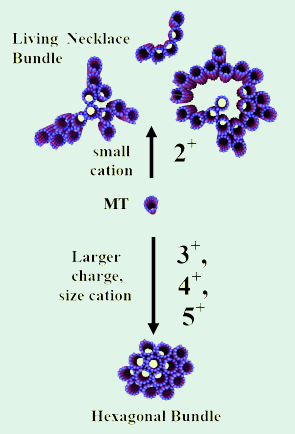
Figure 1. Three dimensional schematics of higher
order nanometer scale assembly of microtubules.
Tri-, tetra-, and pentavalent cations lead to the
formation of hexagonal bundles (left). Small
divalent cations (Ba2+, Ca2+, Sr2+) lead to
the living necklace bundles with linear, branched,
and loop morphologies (right) [8].
Microtubule bundles
MTs are often organized into bundles in vivo. These bundles show a wide variety of internal organizations and sizes. In many instances the biological significance of MT bundle formation is still unclear. For example, the role of MT bundles observed at the hillock region of nerve cells and also that of MTs protruding along the axon are unknown [1]. MT organization in vivo is directed by MAPs [2]. However, even though MAPs can bundle MTs in vitro [3] and in over-expression experiments in vivo [4], the mechanism of bundle formation remains controversial [5]. It is unclear if MAPs crosslink MTs directly, act as spacers between MTs, or facilitate a pre-existing attraction between MTs [5–7]. Furthermore, it is not known how these MAP-mediated microscopic interactions between MTs give rise to the different morphologies of MT bundles observed in vivo.
Microtubule bundles — Our Work (Overview)
We have formed microtubule bundles in vitro using a variety of multivalent cations, inert polymers, and microtubule associated proteins (MAPs). We have characterized the structure of these supramolecular assemblies from the nanoscale to the mesoscale using synchrotron x-ray scattering and diffraction as well as video enhanced DIC, fluorescence and electron microscopy. Even in these simple model systems, the formed bundles vary in degree of branching, persistence length, width, and internal symmetry. These studies have allowed us to probe the nature of MT–MT interactions, and estimate the strength of the lateral bond between protofilaments (and how it is effected by taxol, salt, and MAPs). In addition, we have gained insight into the nature of like charge attraction between rigid polyelectrolytes and helped address the question how microtubules self-assemble into bundles with different structures, as observed in vivo.
The role of MT bundles in nerve cells is not understood and the parameters which determine the bundle size and the nature of MT assembly remain to be elucidated. Our experiments with reconstituted MT and bundle-inducing counterions have revealed two main electrostatically stabilized bundle phases of MTs, which are not predicted by current electrostatic theories of polyelectrolyte bundling. They constitute a step toward understanding how varying microscopic interactions lead to the wide variety of MT bundles observed in vivo [8].
MT bundles — Effect of counterions
It turns out that the charge of the counterion is a key experimental parameter controlling the bundle size and, unexpectedly, the structure and symmetry of the assembly. In addition to providing insight into the fundamental physics of rod-like polyelectrolytes and the general determinants of bundle structure and formation, the control of bundle morphology demonstrated in this system may help to assemble nanostructures for engineering and biomedical applications. The morphologically distinct MT assemblies may be used as templates for miniaturized materials requiring high volume (hexagonal bundles) or high surface area (necklace bundles), with applications in nanotechnology and biotechnology.
The results of our studies of the assembly behavior of multivalent cations and microtubules [8] are summarized in Figure 1. Tightly packed hexagonal bundles with controllable diameters are observed for tri-, tetra-, and pentavalent counterions. Unexpectedly, in the presence of divalent metal cations (Ca2+, Sr2+, Ba2+), we have discovered a new phase, which we refer to as the living necklace bundle phase of MTs. This phase is comprised of dynamical, highly asymmetric self-assembled nematic membranes of MT with linear, branched, and loop topologies and nematic in-plane ordering.

Figure 2. (A) Differential interference contrast (DIC) optical
micrographs of hexagonal microtubule (MT) bundles formed
with 4+ (5 mM spermine) and 2+ (100 mM BaCl2) counter-
ions. (B) (left) Whole mount TEM side view of hexagonal MT
bundles (10 mM spermine) and (right) plastic embedded TEM
cross section. (C) Plastic embedded TEM cross sections of
bundles with 100 mM BaCl2 showing (left) linear, (center)
loop-like and (right) branched morphologies [8].
The mesoscopic structure of MT bundles is shown in video-enhanced DIC images in Figure 2A. The bundles formed in the presence of tri-, tetra-, and pentavalent cations, (such as Spermine4+) appear thick and curved, while bundles formed with small divalent cations (such as Ba2+) are straight. TEM shows that bundles formed with multivalent ions are thick, with MTs tightly packed into a hexagonal array (Figure 2B). A radically different bundle structure is observed when the condensing ions are small, divalent metal cations. TEM shows that these necklace bundles are highly flexible in cross-section, giving rise to topologically distinct linear, branched, and loop morphologies (Figure 2C).
Synchrotron small angle x-ray scattering and diffraction (SAXRD) experiments, performed at the SSRL beamline 4-2 have enabled further insight into the structure of these MT bundles. We have quantitatively modeled the SAXRD data after background subtraction (Figure 3). The MTs are modeled as hollow cylinders, and the tight bundle phase as a collection of MTs packed into a hexagonal lattice. These bundles are finite-size, hexagonal, columnar liquid crystals [9] and the average bundle thickness can be determined from the peak width using Warren’s approximation [10]. As the charge of the condensing ion decreases from 5+, to 4+, to 3+, the MT center-to-center distance increases and the bundle size decreases.
SAXRD scans of bundles assembled with divalent ions display very broad peaks (Figure 3A, "2+"). Our detailed analysis shows that this SAXRD data can be quantitatively modeled as arising from dimers of MTs, confirming that these living bundles are finite size, locally two dimensional membranes with nematic ordering, i.e. they consist of rod-like subunits (MTs) that spontaneously break symmetry by orienting but show only short range positional order. These bundles are an experimental realization of nematic membranes, which have recently been predicted as a new universality class of membrane [11].
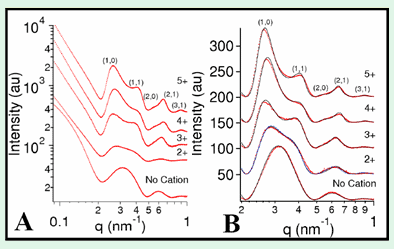
Figure 3. (A) Raw SAXRD scattering data for MTs without added
cation ("No Cation"),
115 mM BaCl2 ("2+"), 15 mM spermidine ("3+"),
5 mM spermine ("4+"),
or 5 mM Penta(L-lysine) ("5+") with hexagonal
bundle peaks indexed.
(B) Data in (A) after background subtraction (dots) with fitted model
scattering curves (lines). Adapted from [8].
[1] Hirokawa, N.: Molecular architecture and dynamics of the neuronal cytoskeleton; In: The neuronal cytoskeleton; R.D. Burgoyne, Ed.; Wiley: New York, 1991; p. 5-74.
[2] MacRae, T. H.: Microtubule organization by cross-linking and bundling proteins. Biochim. Biophys. Acta 1992, 1160, 145-155.
[3] Lida, J.; et al.: The projection domain of MAP4 suppresses the microtubule-bundling activity of the microtubule-binding domain. J. Mol. Biol. 2002, 320, 97-106.
[4] Chen, J.; et al.: Projection domains of MAP2 and Tau determine spacings between microtubules in dendrites and axons. Nature 1992, 360, 674-677.
[5] Chapin, S. J.; Bulinski, J. C.; Gundersen, G.G.: Microtubule bundling in cells. Nature 1991, 349, 24.
[6] Marx, A.; et al.: On the rigidity of the cytoskeleton: are MAPs crosslinkers or spacers of microtubules? Cell. Mol. Biol. 2000, 46, 949-965.
[7] Lee, G.; Brandt, R.: Microtubule-bundling studies revisited: is there a role for MAPs? Trends Cell Biol. 1992, 2, 286-289.
[8] Needleman, D. J.; Ojeda-Lopez, M. A.; Raviv, U.; Miller, H. P.; Wilson, L.; Safinya, C. R.: Higher-order assembly of microtubules by counterions: From hexagonal bundles to living necklaces. Proc. Natl. Acad. Sci. U.S.A. 2004, 101, 16099-16103.![]()
[9] Selinger, J. V.; Bruinsma, R. F.: Phys. Rev. A 1991, 43, 2910-2921.
[10] Warren, B. E.: Phys. Rev. 1941, 59, 693-698.
[11] Xing, X.; Mukhopadhyay, R.; Lubensky, T. C.; Radzihovsky, L.: Phys. Rev. E 2003, 68, 021108.
Cationic Liposome–Microtubule Complexes:
Lipid–Protein Bio-Nanotubes with Open or Closed Ends [1]
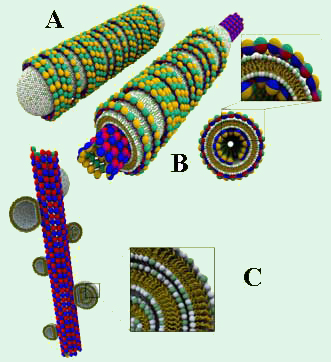
Figure 1. A. Schematic of a lipid protein nanotube (LPN)
with closed ends and lipid caps [1]. B. An LPN with open
ends and a cross section. C. Schematic of the BOR
structure, in which cationic vesicles are absorbed onto
negatively charged MTs. We have explored the structures that form when cationic liposomes are mixed with microtubules (MTs). Using synchrotron small angle X-ray diffraction (performed at the Stanford Synchrotron Radiation Laboratory (SSRL), beam-line 4-2) and transmission electron microscopy (TEM), we have discovered two novel structures [1]. One of these, dubbed lipid–protein–nanotube (LPN), forms when the cationic liposomes spread and coat the MTs and the external lipid layer is decorated by tubulin oligomers (Figure 1 (A,B)). By controlling the cationic lipid/tubulin stoichiometry of the complex, the LPNs can switch between a state with open ends (Figure 1A) and a state with closed ends with lipid caps (Figure 1B). Since the basic governing concepts for this self assembly are universal, this may eventually be exploited for controlled drug encapsulation and release by employing synthetic mimics of LPNs (e.g. replacing microtubules with rigid polyelectrolyte tubes).
For low lipid membrane charge density, we find that the cationic liposomes adsorb onto the MT and appear as "beads on a rod" (BOR; Figure 1C). Interestingly, this straightforward structure was previously hypothesized for cationic lipid–DNA complexes [2], but later experimentally proven to be unstable [3,4]. While the cationic liposome–MT BOR structure is stable enough to be studied, it is a kinetically trapped state which converts to LPNs on longer timescales (several days).
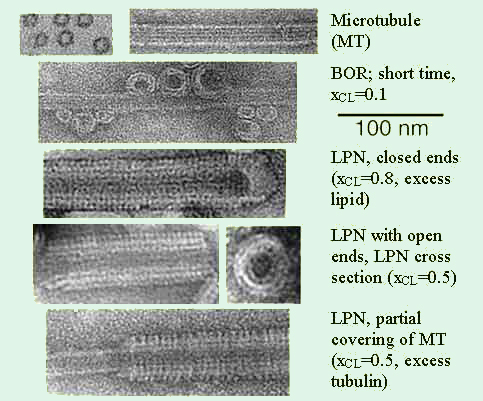
Figure 2. Transmission electron microscopy images of the various states [1]. Whole
mount images of pure MTs as a control, and cationic lipid–MT complexes. The
lipid was a mixture of cationic DOTAP and neutral DOPC with xCL the mole
fraction
of DOTAP.
A set of TEM images covering the structures in the phase diagram of MT–membrane complexes are shown in Figure 1. For a cationic lipid mole fraction of xCL = 0.1, we initially find the BOR structure. For xCL > 0.1, LPNs form immediately upon mixing. The images reveal that LPNs are made up of three layers: Intact MTs are coated by a lipid bilayer (brighter in the images, as the ionic stain avoids the hydrophobic lipid tails), which in turn is coated by tubulin oligomers forming rings or spirals. The oligomer orientation is perpendicular to the internal MT protofilament direction and their density increases with xCL. The three layered LPN structure arises because of the mismatch between the charge densities of MTs and cationic membranes, with the oligomers coating the external lipid monolayer to optimize the electrostatic interactions and counterion release. At a given mole fraction of cationic lipid xCL, it depends on the total amount of added lipid whether the MTs are only partially coated and form LPNs with open ends or LPNs with closed ends with lipid caps, where excess vesicles are attached primarily to the ends of the LPNs.
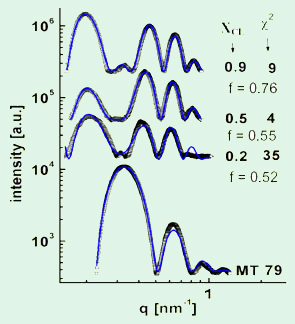
Figure 3. Analysis of the synchrotron small angle
x-ray scattering data [1]. Pictured are radially averaged
scattering intensities of MT-lipid complexes (open
symbols) at various xCL after background subtraction.
The solid blue lines are the fitted scattering profiles.
The fit allowed determining the tubulin oligomer
coverage, f, on the outside of the LPNs.
By performing a detailed analysis of their SAXRD data, we gained quantitative insight into the organization of the complexes. For this purpose, we fitted the SAXRD data (after background subtraction) to a model of the electron density profile of the LPNs. Figure 3 shows examples of data (open squares) and the corresponding fits (blue line). This method allowed us to quantify average coverage in the outer tubulin layer of the LPNs as a function of lipid charge density and lipid/MT charge stoichiometry. Thus, we were able to construct a state diagram for the cationic lipid–MT complexes that furthers our understanding of lipid–polyelectrolyte complexes.
The described results also are of relevance for nonviral gene delivery, since positively charged membranes are commonly used for nonviral drug and gene delivery and introduced into cells [4]. Our results suggest that their association with microtubules, leading to possible changes in their structure and the dynamics in cells, has to be an important consideration in delivery applications using cationic lipids.
[1] Raviv, U.; Needleman, D. J.; Li, Y. L.; Miller, H. P.; Wilson, L.; Safinya, C. R.: Cationic liposome-microtubule complexes: Pathways to the formation of two-state lipid-protein nanotubes with open or closed ends. Proc. Natl. Acad. Sci. U.S.A. 2005, 102, 11167-11172.![]()
[2] Felgner, P. L.; Gadek, T. R.; Holm, M.; Roman, R.; Chan, H. W.; Wenz, M.; Northrop, J. P.; Ringold, G. M.; Danielsen, M.: Proc. Natl. Acad. Sci. U.S.A. 1987, 84, 7413-7417.
[3] Rädler, J. O.; Koltover, I.; Salditt, T.; Safinya, C. R.: Structure of DNA-cationic liposome complexes: DNA intercalation in multilamellar membranes in distinct interhelical packing regimes. Science 1997, 275, 810-814.![]()
[4] Koltover, I.; Salditt, T.; Rädler, J. O.; Safinya, C. R.: An inverted hexagonal phase of cationic liposome-DNA complexes related to DNA release and delivery. Science 1998, 281, 78-81.![]()
Protein Networks of Neurofilaments (NFs) Purified from the Bovine Central Nervous System
Motivation
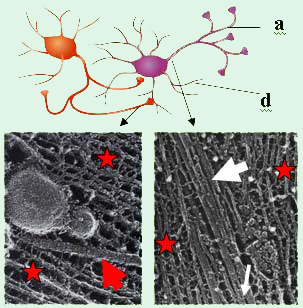
Figure 1. Top: Sketch of two interacting nerve
cells
(a = axon, d = dendrite).
The axon of the cell
on the
left is forming synaptic junctions with the cell
body
and dendrite of the cell on the right.
Lower
Left: Electron micrograph of a
mouse nerve
axon
showing a single microtubule (MT, red arrow)
with
a vesicle
moving along the MT surrounded by
Neurofilaments (NFs, red stars) [1].
Lower Right:
Electron micrograph
of a frog spinal nerve.
A MT
(large white
arrow) embedded within the NF
network
(red stars). MT-associated proteins
(tau
protein,
thin white arrow) noncovalently crosslink
the MTs [2]. The
diameter of an MT is 25 nm.
The axonal cytoskeleton in vertebrate neurons is rich in examples of bundles and networks of neurofilaments, microtubules (MT) and filamentous actin. The nature of these complex structures, their interactions, and the structure–function correlations remains poorly understood. Figure 1 illustrates an example: two interacting nerve cells are shown schematically on the top. The lower left shows an electron micrograph of a longitudinal cut along a nerve cell axon, which exposes the axonal cytoskeleton [1]. A single MT (red arrow) with attached vesicles is visible near the middle. Here, the MT provides a track for the transport of, e.g., vesicles containing precursors of neurotransmitters required for synaptic signal transduction, towards an axon–dendrite or axon–cell body synapse. In the lower right of Figure 1, a section near the initial axon segment close to the cell body is shown in a similar electron micrograph [2]. A finite-size bundle of MTs is clearly visible (large white arrow). MT bundles are thought to be stabilized by microtubule-associated proteins (MAPs; thin white arrow), which noncovalently crosslink neighboring MTs through a combination of electrostatic, hydrogen bonding, and possible hydrophobic interactions. Our ongoing experiments are centered on our desire to understand how cytoskeletal proteins, purified from neurons, assemble into hierarchical structures. The implication of the cytoskeletal structures in many neurodegenerative diseases such as Alzheimer’s disease motivates us to understand the physics of their assembly from purified proteins, reconstituted in vitro. The ultimate goal of this work is to relate structure to biological function. Thus, we are studying nanoscale assembly in model systems comprised of reconstituted neurofilaments (NFs, purified from bovine spinal chord) and microtubule (MT)/microtubule-associated-protein (MAP) complexes. Our recent work on MT/MAP complexes (preprint form) will be described in an update here in the near future.
Neurofilament Architecture
Both the single MT (Fig. 1, lower left) and the MT bundle (Fig. 1, lower right) in the EM images are surrounded by a network of neurofilaments (NFs, red stars), which play a major role in the structural stability of the axon of vertebrate neurons. As schematically shown in Figure 2A, neurofilaments are self-assembled heteropolymers, consisting of three proteins with distinct molecular weight (MW): NF-L(ow MW), NF-M(edium MW), and NF-H(igh MW). These components are readily resolved by SDS PAGE as seen in Figure 2B.
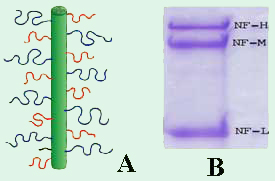
Figure 2. (A) Schematic of a neurofilament (NF)
showing the sidearms of NF-H (blue), NF-M (red),
and NF-L (black) fractions which contain 600, 500
and 150 amino acid residues, respectively. (B)
SDS PAGE Gel (10% polyacrylamide) of purified
NFs, showing the three fractions
(NF-L: MW = 60
kDa; NF-M: MW = 100 kDa;
NF-H: MW = 115
kDa) [7].
Upon reconstitution, purified NF fractions spontaneously assemble to form a core filament with an overall negative charge. From this core, sidearms of varying length that are rich in carboxylic acid-bearing amino acids extend (Figure 2A). The electron micrographs in Figure 1 show the crossbridging between neighboring neurofilaments resulting from sidearm interactions. A known hallmark of motor neuron diseases such as amyotrophic lateral sclerosis is the disruption of the NF-network due to incorrect sidearm interactions [3].
Neurofilament results
The precise nature of short-range and long-range interactions between NFs resulting from sidearm interactions remains controversial, primarily due to the lack of quantitative data. We purify NFs from bovine spinal cord [4–6], separating the three NF proteins by chromatographic techniques. Recent experiments on NFs reconstituted from their components show that the NFs form a liquid crystalline gel phase. This is clearly visible in polarized microscopy images such as the one shown in Figure 3. The texture here is characteristic of a nematic liquid crystalline network with long-range orientational order within the filamentous network. Small angle x-ray scattering data from reconstituted NF mixtures shows promise of being able to reveal the role of sidearm interactions in controlling inter-filament interactions.
In our most recent work [8], we reassembled NFs in vitro from varying weight ratios of the subunit proteins, purified from bovine spinal cord, to form homopolymers of NF-L or filaments composed of NF-L and NF-M (NF-LM), NF-L and NF-H (NF-LH), or all three subunits (NF-LMH). At high protein concentrations, NFs were found to align to form a nematic liquid crystalline gel with a well-defined spacing determined by synchrotron small angle x-ray scattering. Near physiological conditions (86 mM monovalent salt and pH 6.8), NFLM networks with a high NF-M grafting density favor nematic ordering whereas filaments composed of NF-LH transition to an isotropic gel at low protein concentrations as a function of increasing mole fraction of NF-H subunits. The interfilament distance decreases with NF-M grafting density, opposite of the trend seen with NF-LH networks. This suggests a competition between the more attractive NF-M sidearms, forming a compact aligned nematic gel, and the repulsive NF-H sidearms, favoring a more expansive isotropic gel, at 86 mM monovalent salt. These interactions are highly salt dependent and the nematic gel phase is stabilized with increasing monovalent salt.
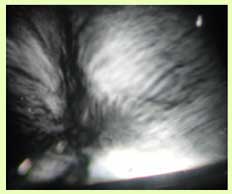
Figure 3. A typical reconstituted NF
mixture, viewed between crossed polarizers,
shows a nematic-like liquid crystalline texture [7].
[1] Hirokawa, N.: Trends Cell Biol. 1996, 6, 135-141.
[2] Peters, A.; Palay, S. L.; Webster, H.: The Fine Structure of the Nervous System; 3rd edition, 1991; Oxford, New York.
[3] Miller, C. C. J.; Ackerley, S.; Brownlees, J.; Grierson, A. J.; Jacobsen, N. J. O.; Thornhill, P.: Cell. Mol. Life Sci. 2002, 59, 323-330.
[4] Leterrier, J. F.; Eyer, J.: Biochem. J. 1987, 245, 93-101.
[5] Leterrier, J. F.; Käs, J.; Hartwig, J.; Vegners, R.; Janmey, P. A.: J. Biol. Chem. 1996, 271, 15687-15694.
[6] R. K. Liem, 1986, 134, 384-388.
[7] Needleman, D. J.; Jones, J. B.; Raviv, U.; Ojeda-Lopez, M. A.; Miller, H. P.; Li, Y.; Wilson, L.; Safinya, C. R.: Supramolecular assembly of biological molecules purified from bovine nerve cells: from microtubule bundles and necklaces to neurofilament networks. J. Phys.: Condens. Matter 2005, 17, S3225-S3230.![]()
[8] Jones, J. B.; Safinya, C. R.: Interplay between Liquid Crystalline and Isotropic Gels in Self-Assembled Neurofilament Networks. Biophys. J. 2008, 95, 823-825.![]()
Cytoskeletal Proteins in Microchannels