Introduction: why Microchannels?
In order to be relevant for our understanding of biology, studies of protein self-assemblies preferably should be performed under physiological conditions. This can be challenging, since biological self-assembled systems formed under physiological conditions in vitro can be very delicate structures and, therefore, extremely difficult to manipulate. Thus, the study of self-assembled protein systems and their structure–function relationships will require new techniques for the confinement and manipulation of specific molecular assemblies.
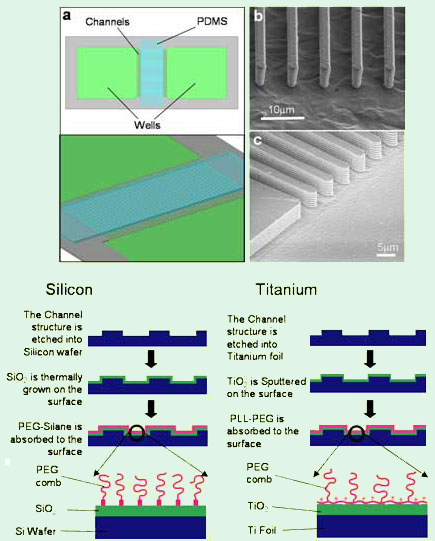
Figure 1. Left: Schematic depiction of the microfluidic device (a) and
SEM images of the channel
ends in (b) titanium and (c) silicon.
Right:
Schematic depiction of the fabrication process of Si
and
Ti microchannels.
Structures such as cytoskeletal protein bundles or lipid-DNA complexes will form only under specific conditions and cannot be crystallized for detailed structural studies. These self-assembled structures tend to be composed of weakly ordered large molecules, which means that X-ray scattering will be very weak. Therefore, typical X-ray scattering samples in such systems consist of a concentrated pellet of material, produced by centrifugation. The production of such a sample generally requires a large amount of protein. However, many proteins are costly and difficult to purify, thus limiting the amounts of protein available for study. In addition, due to their inherent properties (fragility etc.), most of the X-ray scattering work on such systems to date has been performed on unoriented (powder) samples. Whilst such work is able to provide some information on molecular spacings in a complex, much more detailed information could be obtained from aligned samples. Alignment also enhances the signal to noise ratio of X-ray scattering experiments, allowing samples of lower concentration to be investigated. Therefore, an ideal device would be able to utilize dilute solutions of fragile samples and align them, allowing investigations of the material to yield a maximum of structural information. This provides the motivation for our studies on microchannels
Microchannels in Titanium and Silicon for Studies of Aligned Protein Self-Assemblies
We have developed a technique which makes it possible to investigate the structural and mechanical properties of self-assembled protein systems by confinement in a surface-modified microfluidic device, produced via a high aspect ratio etch process. This technique allows self-assembly to occur under defined conditions whilst minimizing external influences, e.g. shear forces or attraction to a surface. By forming actin bundles with α-actinin or mictrotubules in a narrow channel, where the width of the channel is smaller that the persistence length of the filamentous structure, we have produced highly aligned protein self-assemblies. Biomolecular self-assembly can be investigated in a controlled fashion under different molecular concentration gradients and conditions along a channel length. At the same time, we aim to carry out X-ray scattering studies on very small volumes of material. This technique has been demonstrated on a silicon device [1], to give a highly aligned X-ray diffraction pattern for the α-actinin/actin bundle system.

Figure 2. Confocal microscopy images of F-actin bundles
formed in the presence
of α-actinin at a molar ratio of 1:5
α-actinin/G-actin. Left: Bundles formed without
confinement
(a) and in 10 mm silicon microchannels (b,c,d). (d) shows
a close-up
as marked in (c). Right: F-actin bundles formed
in the presence of α-actinin in 20μm titanium microchannels
and schematic depiction of α-actinin–actin bundles [3].
In addition to silicon, we have recently used titanium (Ti) as the device material. Titanium is a very interesting material for microdevice fabrication. The metal is biocompatible and already utilized widely in medical implants. The titanium deep etch process is relatively new and provides us with a material which is extremely robust. Devices can be thinned to as little as 5 μm whilst retaining their structural integrity and patterned with small features comparable to those commonly produced in silicon. The surface of Ti devices can be modified with PEG chains to resist protein absorption. For X-ray scattering applications, we have found that the polycrystalline titanium substrates show significant advantages over silicon wafers in both background scattering and robustness.
Figure 1 shows a schematic and SEM images of actual microfluidic devices on the left, while the fabrication processes for Si and Ti are outlined on the right.
Using shallow channels, the molecular dynamics of actin filaments and bundles, microtubules, and other filamentous proteins can be observed in a controlled way via fluorescence microscopy. Chemical gradients can be applied along the channels and electric field conditions controlled by micropatterning the device with insulating and conducting surfaces.
The following figures show a few examples of biomacromolecules under confinement in our microfluidic devices. Notably, Figure 2 shows examples of 10-20 μm wide channels filled with α-actinin/actin bundles. Thick individual bundles can clearly be resolved. Experiments using silicon (Figure 2 b-d) and titanium microchannels (Figure 2 e-g) yielded similar results. In (b), the curvature of the bundle structure in the channel can easily be observed, showing a marked difference from the unconfined bundles (a). Many branch points can also be observed. It is interesting to note that the bundles appear to have formed in response to a gradient, as highlighted by Figure 2 (g). Thick bundles have formed at the channel end close to the α-actinin reservoir, apparently getting thinner towards the actin reservoir. This indicates that bundle thickness is a function of α-actinin/actin ratio in solution, a result also observed in bulk samples [2,3].
Another interesting protein system to study via alignment are microtubules and their associated proteins (MAPs). As with the linker proteins attaching to the actin filaments, it should also be possible to study the arrangement of MAPs along the microtubule if we can align this system for X-ray scattering in the same way. Microtubules were polymerized in titanium microchannels and the results can be seen in Figure 3. A solution of 4 mg/ml tubulin in a buffer suitable for polymerization was introduced to channels of different thickness on ice, and then incubated at 37 °C to induce polymerization.
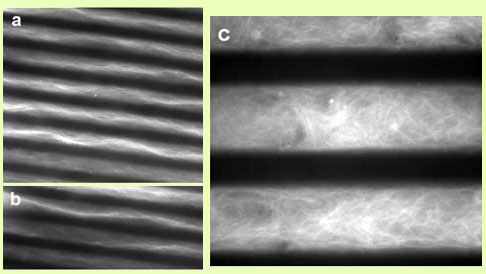
Figure 3. Fluorescence microscopy images of microtubules polymerized in-situ
in 5 μm wide (a and b)
and 20 μm wide (c) titanium channels.
As shown in Figure 3, the microtubules polymerized into an aligned state in 5 μm wide channels, while very little alignment was observed in 20 μm channels. Microtubules are fairly stiff polymers, with a persistence length of ~20 μm. Alignment is expected for channel widths smaller than the persistence length, as observed in our experiments.
[1] Bouxsein, N. F.; Hirst, L. S.; Li, Y. L.; Safinya, C. R.; Abu Samah, Z.; MacDonald, N. C.; Pynn, R.: Alignment of filamentous proteins and associated molecules through confinement in microchannels. Appl. Phys. Lett. 2004, 85, 5775-5777. ![]()
[2] Hirst, L. S.; Safinya, C. R.: Skin layer at the actin-gel surface: Quenched protein membranes form flat, crumpled, and tubular morphologies. Phys. Rev. Lett. 2004, 93, 018101. ![]()
[3] Pelletier, O.; Pokidysheva, E.; Hirst, L. S.; Bouxsein, N.; Li, Y.; Safinya, C. R.: Structure of actin cross-linked with alpha-actinin: A network of bundles. Phys. Rev. Lett. 2003, 91, 148102. ![]()
Confinement of Liquid Crystals in Microchannels
For information on this work see:
Choi, M. C.; Pfohl, T.; Wen, Z. Y.; Li, Y. L.; Kim, M. W.; Israelachvili, J. N.; Safinya, C. R.: Ordered patterns of liquid crystal toroidal defects by microchannel confinement. Proc. Natl. Acad. Sci. U.S.A. 2004, 101, 17340-17344. ![]()